Conversion rate analysis
Pavel Reich
29-Jun-2020 08:35 CEST
Let’s simulate user visits with 4 different browser versions, 1 bad and 3 good. Main languages of users are de/fr/en and they don’t make a significant difference on conversion. The version does and we want to detect it using Pairwise Wilcoxon Rank Sum Test to identify pairs with different conversion rate and Chi-squared Test to identify significant variables (it’s version, not language).
library(ggplot2)
generateVisitors=function(version, lang,cnt, success_rte) {
data.frame(version=version, lang=lang,success=rbinom(cnt,1,rep(success_rte,cnt)))
}
generateVisitorData=function(bad_version_visitors, total) {
df=data.frame()
df=rbind(df, generateVisitors("good-version.1", "de",round(total*0.45),0.12))
df=rbind(df, generateVisitors("bad-version.2", "en",bad_version_visitors,0))
df=rbind(df, generateVisitors("good-version.3", "en",round(total*0.34),0.15))
df=rbind(df, generateVisitors("good-version.4", "fr",round(total*0.11),0.12))
df
}
df=generateVisitorData(70,870)
df[sample(1:nrow(df), size = 20),]## version lang success
## 17 good-version.1 de 0
## 508 good-version.3 en 0
## 657 good-version.3 en 0
## 357 good-version.1 de 0
## 529 good-version.3 en 0
## 744 good-version.3 en 1
## 633 good-version.3 en 0
## 439 bad-version.2 en 0
## 420 bad-version.2 en 0
## 32 good-version.1 de 0
## 455 bad-version.2 en 0
## 246 good-version.1 de 0
## 198 good-version.1 de 0
## 384 good-version.1 de 0
## 823 good-version.4 fr 0
## 403 bad-version.2 en 0
## 239 good-version.1 de 0
## 451 bad-version.2 en 0
## 335 good-version.1 de 1
## 178 good-version.1 de 0aggregate(success ~ lang, df, mean)## lang success
## 1 de 0.1352041
## 2 en 0.1338798
## 3 fr 0.1458333aggregate(success ~ version, df, mean)## version success
## 1 good-version.1 0.1352041
## 2 bad-version.2 0.0000000
## 3 good-version.3 0.1655405
## 4 good-version.4 0.1458333pairwise.wilcox.test(df$success, df$version)##
## Pairwise comparisons using Wilcoxon rank sum test
##
## data: df$success and df$version
##
## good-version.1 bad-version.2 good-version.3
## bad-version.2 0.0044 - -
## good-version.3 0.8042 0.0016 -
## good-version.4 1.0000 0.0044 1.0000
##
## P value adjustment method: holmpairwise.wilcox.test(df$success, df$lang)##
## Pairwise comparisons using Wilcoxon rank sum test
##
## data: df$success and df$lang
##
## de en
## en 1 -
## fr 1 1
##
## P value adjustment method: holmchisq.test(table(df$version,df$success))##
## Pearson's Chi-squared test
##
## data: table(df$version, df$success)
## X-squared = 13.312, df = 3, p-value = 0.004009chisq.test(table(df$lang,df$success))##
## Pearson's Chi-squared test
##
## data: table(df$lang, df$success)
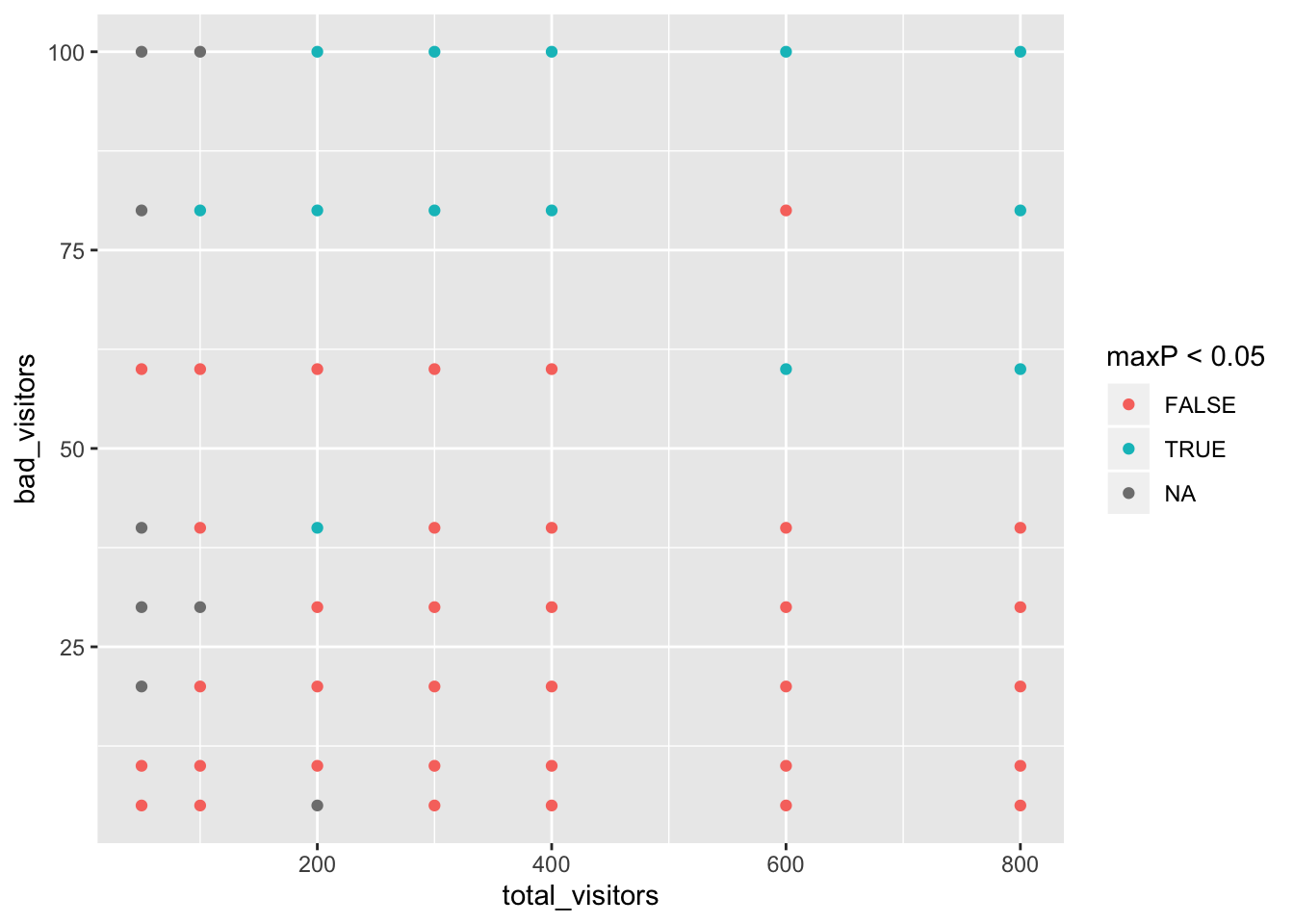
## X-squared = 0.095007, df = 2, p-value = 0.9536Now let’s see how many visitors in total and with a bad version we need to detect problems with max p-value of 0.05 across pairs badVersion and goodVersion1,3,4.
ret=data.frame()
for (bad_visitors in c(5,10,20,30,40,60,80,100)) {
for (total_visitors in c(50,100,200,300,400,600,800)) {
df=generateVisitorData(bad_visitors, total_visitors)
p=pairwise.wilcox.test(df$success, df$version)
ret=rbind(ret, data.frame(bad_visitors=bad_visitors, total_visitors=total_visitors, maxP=max(c(p$p.value[1,1], p$p.value[2:3,2]))))
}
}
ggplot(ret, aes(x=total_visitors, y=bad_visitors, colour=maxP<0.05))+geom_point()